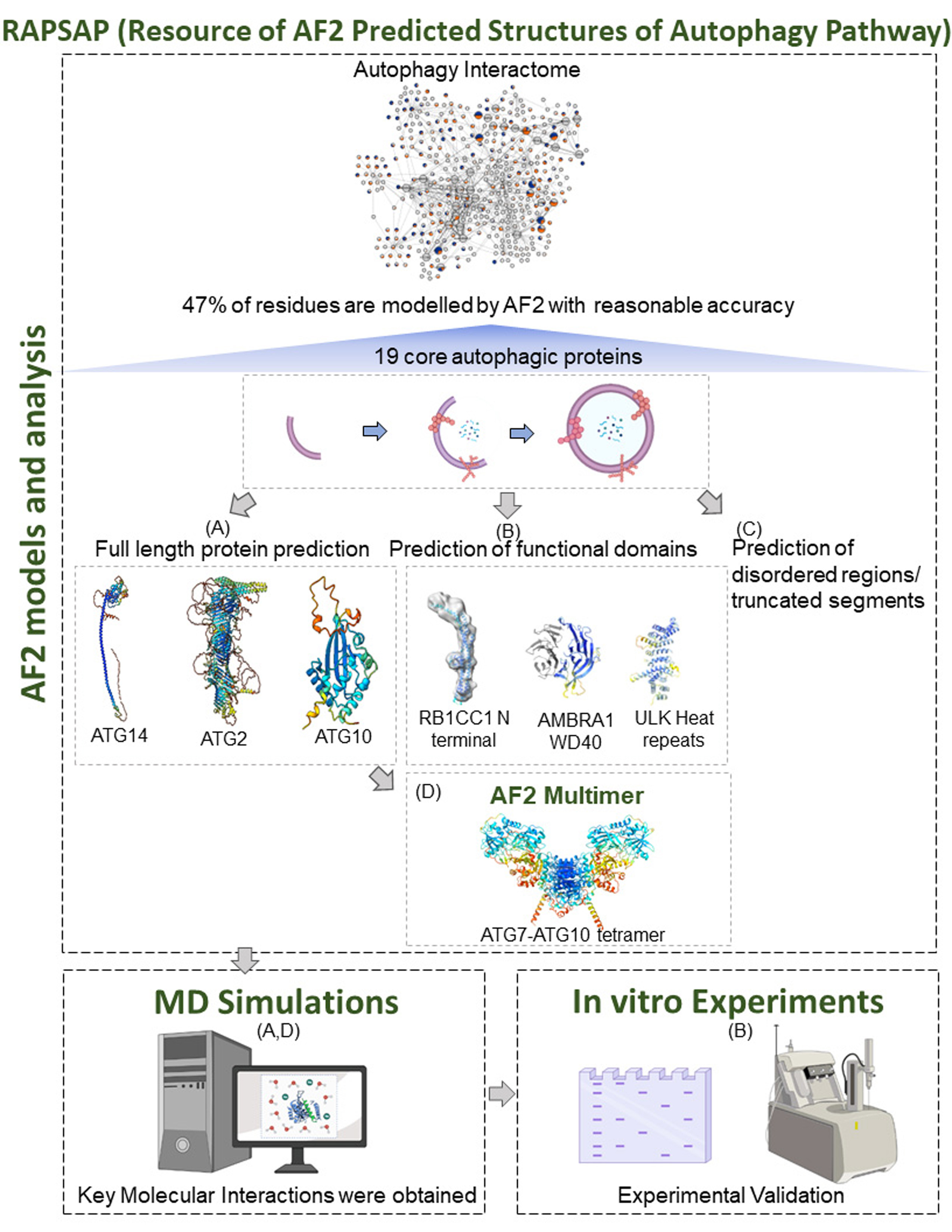
The RAPSAP resource can be broadly categorised to host two set of information, first AF2 models and analysis, and secondly MD simulation related data. As a meta-data approach, we gathered predicted models by AF2 of core autophagy proteins and its interactors (~ 416 proteins). The models are then analysed on the basis of pLDDT score and the structured and disordered ratio. The webpage AF models and analysis reports precisely average pLDDT score, BLASTp results, pLDDT score on the predicted segment, and intrinsically disordered analysis based on IUPred2A. A list of non-IDRs with high pLDDT scores are provided to enable the readers to identify functionally important regions predicted by AF2.
Secondly, ensemble of high confidence structures of three core autophagic proteins (ATG2, ATG10, and ATG14) was generated using micro second MD simulations. In addition, AF2 multimeric complex models of ATG7-ATG10 along with structural ensemble from simulations, is made freely available to the readers. The key molecular interactions crucial to the complex formation was experimentally validated using mutagenesis and Isothermal Titration Calorimetric experiments.
Framework of RAPSAP
